New Optimizer
Kirstine.jl comes with a simple particle swarm optimizer. This document describes how to implement a custom particle-based optimizer that seamlessly plugs into the rest of the package, such that it can be used with any Kirstine.ProblemSolvingStrategy.
Our example we will be a simplified version of the Imperialist-Competitive Algorithm (ICA) from the ICAOD R package. ICA is based on the following metaphor: a set of points represents countries, some of which are imperialists and some of which are colonies. An imperialist together with its associated colonies forms an empire. Initially, a given number of countries are designated as imperialists and colonies are assigned randomly. After that, the following steps are repeated:
- Assimilation: all colonies move a bit towards their imperialist.
- Revolution: some colonies move around randomly. The revolution rate decreases exponentially over time.
- Swap: a colony that has a better objective function value than its imperialist becomes the new imperialist of the empire.
- Unite: if the distance between two imperialists falls below a certain threshold, their empires are merged.
- Competition: in some iterations, stronger empires can try to steal a colony from the weakest empire. Strength of an empire is a weighted average of the objective functions values of its countries. If an empire has only zero or one colonies left, it is merged into the strongest empire.
Abstract Points
Before we get to the implementation details of ICA, we first need to understand how the optimization code in Kirstine.jl is structured. Particle-based optimizers typically expect to work with vectors from $\Reals^n$, as they need to perform the following operations on $x,y∈\Reals^n$ and $α∈\Reals$:
- $(x, y) ↦ x - y$: find the directional vector for moving from $y$ to $x$
- $(x, y) ↦ x + y$: update the position
- $(α, x) ↦ α x$: scale
- $(x, y) ↦ (x_1 y_1, …, x_n y_n)$: multiply elementwise
Of course, the set of DesignMeasures is not a vector space. But we can map a design measure
\[\DesignMeasure = \sum_{\IndexDesignPoint=1}^\NumDesignPoints \DesignWeight_{\IndexDesignPoint} \DiracDist(\DesignPoint_{\IndexDesignPoint})\]
on $\DesignRegion ⊂ \Reals^{\DimDesignRegion}$ to a vector in $\Reals^{\NumDesignPoints(\DimDesignRegion + 1) - 1}$ by
\[f(\DesignMeasure) = (\DesignPoint_1,…,\DesignPoint_{\NumDesignPoints}, \DesignWeight_1,…,\DesignWeight_{\NumDesignPoints-1})\]
and let the optimizer work on the representations $f(\DesignMeasure)$. Note that the final weight $\DesignWeight_{\NumDesignPoints}$ is not mapped, since it is implicitly known from the first $\NumDesignPoints - 1$ weights. A problem now is that in general there is no design measure
\[\text{“ } f^{-1}(f(\DesignMeasure_1) - f(\DesignMeasure_2)) \text{ ”}\]
since it could have negative weights or points outside $\DesignRegion$. Hence we need a different kind of object to represent these differences. For this, we used discrete signed measures. We denote the implied subtraction, addition, and multiplication operators by $⊖, ⊕, ⊙$. When adding a signed measure $\SignedMeasure$ to a design measure $\DesignMeasure$, we must take care that the weights and design points of the result $\DesignMeasure ⊕ \SignedMeasure$ are valid for a design measure.
Note that the set of signed measures is a vector space, where
\[\DesignMeasure_1 - \DesignMeasure_2 = \sum_{\IndexDesignPoint=1}^\NumDesignPoints ( \DesignWeight_{1,\IndexDesignPoint} \DiracDist(\DesignPoint_{1,\IndexDesignPoint}) - \DesignWeight_{2,\IndexDesignPoint} \DiracDist(\DesignPoint_{2,\IndexDesignPoint}) )\]
However, for optimal design purposes we want the number of design points to remain constant and use the difference
\[\DesignMeasure_1 ⊖ \DesignMeasure_2 = \sum_{\IndexDesignPoint=1}^\NumDesignPoints (\DesignWeight_{1,\IndexDesignPoint} - \DesignWeight_{2,\IndexDesignPoint}) \DiracDist(\DesignPoint_{1,\IndexDesignPoint} - \DesignPoint_{2,\IndexDesignPoint})\]
as implied by $f^{-1}$.
Now, from a software engineering point of view, an Kirstine.Optimizer should not have to know anything about the nature of the points it works with. It only sees objects of a type T <: AbstractPoint, and a corresponding type U <: AbstractPointDifference that can store a representation of the difference between two Ts. This separates the optimization logic from the details of how the Ts should pretend to be vectors. The latter behavior is abstracted away into the following methods, shown here for T = DesignMeasure and U = SignedMeasure:
ap_rand!(d::DesignMeasure, c::DesignConstraints): randomizedin place.ap_copy!(to::DesignMeasure, from::DesignMeasure): in-place assignment. Afterwards,toandfromdo not share memory.ap_diff!(v::SignedMeasure, p::DesignMeasure, q::DesignMeasure): compute $v = p ⊖ q$.ap_as_difference(p::DesignMeasure): return theSignedMeasure$v$ such that $f(v) = f(p)$.ap_rand!(v::SignedMeasure, lb::Real, ub::Real): randomize elements ofvin place from a uniform distribution on the interval $[lb, ub]$.ap_mul!(v1::SignedMeasure, v2::SignedMeasure): in-place elementwise multiplication ofv1andv2.ap_mul!(v::SignedMeasure, a::Real): in-place multiplication of each element ofvbya.ap_add!(v1::SignedMeasure, v2::SignedMeasure): update $v_1$ to $v_1 ⊕ v_2$.ap_add!(p::DesignMeasure, v::SignedMeasure, c::DesignConstraints): try to move from $p$ to $p ⊕ v$. If this would result in points outside the design region or invalid weights, move as far as possible in the direction of $v$.
The DesignConstraints <: AbstractConstraints object has the following fields:
dr <: Kirstine.DesignRegiontheKirstine.DesignRegionof the current design problemfixw::Vector{Bool}true elements correspond to design weights that should not movefixp::Vector{Bool}true elements correspond to design points that should not move
When you implement a general Kirstine.Optimizer, you will not need to use this information since it is specific to the DesignMeasure subtype of AbstractPoint.
Implementation
The internal optimization interface consists of functions that operate on objects of a type O <: Kirstine.Optimizer, which holds the algorithm's parameters, and a type S <: OptimizerState which holds the state of the optimizer in the current iteration. For O = Ica and S = IcaState we must implement the following methods:
iterations(optimizer::Ica): accessor for the number of iterations that ICA should runmaximizer(state::IcaState): accessor for the best candidate seen so farmaximum(state::IcaState): accessor for the best objective value seen so farn_eval_f(state::IcaState): accessor for the cumulative number of evaluations of the objective functionn_eval_g(state::IcaState): accessor for the cumulative number of evaluations of the gradient of the objective function (always 0 since ICA does not use gradients)optimizer_state(f, o::Ica, prototypes::Vector{AbstractPoint}, constraints::AbstractConstraints): constructor of anIcaStatefor the objective functionfand parameterso. It should use the givenprototypesfor initialization and take theconstraintsinto account for randomization. Setting up the state is counted as the first iteration of the algorithm.has_converged(state::IcaState, optimizer::Ica): return true if the algorithm has converged already. This is used for early stopping. It is useful for algorithms that have convergence guarantees. Since ICA has no such guarantees, the algorithm never converges (in this strict sense) by definition.tick!(f, state::IcaState, optimizer::Ica, constraints::AbstractConstraints): update the optimizers state once. This function is called internallyiterations() - 1times. The initial state is considered to be the first iteration.
The Kirstine.Optimizer type has two abstract subtypes: A Kirstine.ZerothOrderOptimizer only works with the objective function itself, while a Kirstine.FirstOrderOptimizer also requires its gradient. ICA is of order zero. If you want to implement a first-order optimizer, you need versions of optimizer_state((f, g!), ...) and of tick!((f, g!), ...) that additionally take a mutating gradient g!(g::AbstractVector, x::AbstractPoint) -> g.
using Kirstine
using Statistics: mean
struct Ica <: Kirstine.ZerothOrderOptimizer
iterations::Int64
num_countries::Int64
num_initial_empires::Int64
revolution_rate::Float64 # proportion of colonies where revolution happens
damp::Float64 # damping factor for revolution rate
zeta::Float64 # mean colony weight for power
beta::Float64 # assimilation speed
competition_pressure::Float64 # probability that competition occurs
uniting_threshold::Float64 # min relative distance for imperialists
function Ica(i::Integer, nc::Integer, nie::Integer, rr, d, z, b, cp, ut)
if i <= 0
error("number of iterations must be positive")
end
if d <= 0 || d >= 1
error("damping factor must be in (0, 1)")
end
if cp < 0 || cp > 1
error("competition pressure must be in [0, 1]")
end
if rr < 0 || rr > 1
error("revolution rate must be in [0, 1]")
end
if nie > nc ÷ 2
error("at most half of the countries can be imperialists")
end
return new(i, nc, nie, rr, d, z, b, cp, ut)
end
end
# keyword constructor
function Ica(;
iterations,
num_countries,
num_initial_empires,
revolution_rate,
damp,
zeta,
beta,
competition_pressure,
uniting_threshold,
)
return Ica(
iterations,
num_countries,
num_initial_empires,
revolution_rate,
damp,
zeta,
beta,
competition_pressure,
uniting_threshold,
)
end
Kirstine.iterations(optimizer::Ica) = optimizer.iterations
mutable struct IcaState{T,U} <: Kirstine.OptimizerState{T,U}
country::Vector{T} # all countries
val::Vector{Float64} # function values for all countries
imperialist::Vector{Int64} # contry index for each imperialist
empire::Vector{Vector{Int64}} # country indices for colonies of each empire
power::Vector{Float64} # power of an of empire
rand::U # tmp for uniform random number for assimilation update
diff::U # tmp for difference imperialist - colony
revolution_rate::Float64 # current revolution rate
n_eval::Int64 # cumulative number of objective evaluations
end
Kirstine.maximizer(state::IcaState) = state.country[argmax(state.val[state.imperialist])]
Kirstine.maximum(state::IcaState) = maximum(state.val[state.imperialist])
Kirstine.n_eval_f(state::IcaState) = state.n_eval
Kirstine.n_eval_g(state::IcaState) = 0
function Kirstine.optimizer_state(
f,
o::Ica,
prototypes::AbstractVector{<:Kirstine.AbstractPoint},
constraints::Kirstine.AbstractConstraints,
)
# initialize countries at prototypes, randomize the rest
if o.num_countries < length(prototypes)
error("num_countries must be at least as large as number of prototype particles")
end
cnt_given = deepcopy(prototypes)
n_random = o.num_countries - length(prototypes)
cnt_random = [deepcopy(prototypes[1]) for _ in 1:n_random]
for k in 1:n_random
Kirstine.ap_rand!(cnt_random[k], constraints)
end
country = [cnt_given; cnt_random]
# get the matching difference type for the concrete type of country[1]
zero_diff = Kirstine.ap_as_difference(country[1])
Kirstine.ap_diff!(zero_diff, country[1], country[1]) # initialize at 0
# allocate vectors for the state object
val = fill(NaN, o.num_countries)
imp = fill(0, o.num_initial_empires)
emp = [[0] for _ in 1:(o.num_initial_empires)] # arbitrary invalid index
tc = fill(NaN, o.num_initial_empires)
rnd = deepcopy(zero_diff)
diff = deepcopy(zero_diff)
# instantiate a state object and initialize it properly
state = IcaState(country, val, imp, emp, tc, rnd, diff, o.revolution_rate, 0)
ica_evaluate_objective!(f, state)
num_colonies = o.num_countries - o.num_initial_empires
state.imperialist .= sortperm(state.val, rev = true)[1:(o.num_initial_empires)]
# there is at least one country per empire, with additional ones depending on power
empsize = fill(1, o.num_initial_empires)
min_val = minimum(state.val[state.imperialist])
imperialist_power = (0.3 * sign(min_val) - 1) .* min_val .+ state.val[state.imperialist]
imperialist_power ./= sum(imperialist_power)
# we use the apportionment procedure to make sure empire sizes add up
empsize .+= apportion(imperialist_power, num_colonies - o.num_initial_empires)
# partition the remaining countries according to empire sizes
colony_indices = setdiff(1:(o.num_countries), state.imperialist)
for k in 1:(o.num_initial_empires)
# consume colony index list in empsize-sized chunks
state.empire[k] = colony_indices[1:empsize[k]]
colony_indices = colony_indices[(empsize[k] + 1):end]
end
ica_compute_power!(state, o)
@debug "INIT" state.empire
return state
end
Kirstine.has_converged(state::IcaState, optimizer::Ica) = false
function Kirstine.tick!(
f,
state::IcaState,
optimizer::Ica,
constraints::Kirstine.AbstractConstraints,
)
ica_assimilate!(state, optimizer, constraints)
ica_revolt!(state, optimizer, constraints)
ica_evaluate_objective!(f, state)
ica_swap!(state)
ica_unite!(state, optimizer)
ica_compute_power!(state, optimizer)
ica_compete!(state, optimizer)
end
# For each empire k and colony j,
# move col_{k,j} → col_{k,j} + r .* (imp_{k} - col_{k,j})
# where r is a vector of uniform random numbers on [0, β].
function ica_assimilate!(
state::IcaState,
optimizer::Ica,
constraints::Kirstine.AbstractConstraints,
)
for k in 1:length(state.empire)
imp = state.imperialist[k]
for j in 1:length(state.empire[k])
col = state.empire[k][j]
Kirstine.ap_rand!(state.rand, 0, optimizer.beta)
Kirstine.ap_diff!(state.diff, state.country[imp], state.country[col])
Kirstine.ap_mul!(state.diff, state.rand)
Kirstine.ap_add!(state.country[col], state.diff, constraints)
end
end
return state
end
# Euclidean distance between two DesignMeasures when interpreted as the associated vectors.
# Note that we ignore the implicitly known final weight.
function ap_dist(p::DesignMeasure, q::DesignMeasure)
# we iterate through the scalar elements of the matrix,
# hence we don't use the points() accessor!
ssq = mapreduce((x, y) -> (x - y)^2, +, p.points, q.points)
for k in 1:(length(p.weights) - 1)
ssq += (p.weights[k] - q.weights[k])^2
end
return sqrt(ssq)
end
# For each empire, move a proportion of colonies around randomly. The proportion equals the
# current revolution rate. The maximal distance is four times the distance between colony
# and imperialist.
# A colony col_{k,j} where a revolution happens is moved to
# col_{k,j} + β .* r .* (imp_{k} - col_{k,j})
# where r is a vector of uniform random numbers on [-β/2, β/2]
# I.e. the difference to assimilation is that only some colonies are affected,
# and that they can also move _away_ from their imperialist.
function ica_revolt!(
state::IcaState,
optimizer::Ica,
constraints::Kirstine.AbstractConstraints,
)
state.revolution_rate *= optimizer.damp
for k in 1:length(state.empire)
num_col = length(state.empire[k])
num_rev = Int64.(floor(state.revolution_rate * num_col))
imp = state.imperialist[k]
if num_rev > 0
rev_idx = randperm(num_col)[1:num_rev]
for j in rev_idx
col = state.empire[k][j]
dist = ap_dist(state.country[imp], state.country[col])
Kirstine.ap_rand!(state.rand, -optimizer.beta / 2, optimizer.beta / 2)
Kirstine.ap_add!(state.country[col], state.diff, constraints)
end
end
end
return state
end
function ica_evaluate_objective!(f, state::IcaState)
for i in 1:length(state.country)
state.val[i] = f(state.country[i])
end
state.n_eval += length(state.country)
return state
end
# iterate through empires and swap colony with imperialist if the colony has a better objective value
function ica_swap!(state::IcaState)
for k in 1:length(state.empire)
for j in 1:length(state.empire[k])
country_imp = state.imperialist[k]
country_col = state.empire[k][j]
val_imp = state.val[country_imp]
val_col = state.val[country_col]
if val_col > val_imp
@debug "SWAP colony $j (country $country_col) with imperialist $k (country $country_imp)"
state.imperialist[k] = country_col
state.empire[k][j] = country_imp
end
end
end
return state
end
# loop through pairs of empires and merge any where the imperialists are less than `uniting_threshold` apart
function ica_unite!(state::IcaState, optimizer::Ica)
k = 1
while k < length(state.empire) # note: the length can change in the loop body
l = k + 1
while l <= length(state.empire)
imp_k = state.imperialist[k]
imp_l = state.imperialist[l]
if ap_dist(state.country[imp_k], state.country[imp_l]) <=
optimizer.uniting_threshold
idx_src, idx_dst = state.val[imp_k] > state.val[imp_l] ? (l, k) : (k, l)
@debug "UNITE empire $idx_src with empire $idx_dst"
append!(state.empire[idx_dst], state.empire[idx_src])
push!(state.empire[idx_dst], state.imperialist[idx_src])
deleteat!(state.empire, idx_src)
deleteat!(state.power, idx_src)
deleteat!(state.imperialist, idx_src)
end
l += 1
end
k += 1
end
return state
end
# an empires power is the weighted mean of the objective functions's values at its
# imperialist and its colonies, shifted such that the minimum is zero
function ica_compute_power!(state::IcaState, optimizer::Ica)
for k in 1:length(state.empire)
state.power[k] =
state.val[state.imperialist[k]] +
optimizer.zeta * mean(state.val[state.empire[k]])
end
idx_finite = .!isinf.(state.power)
state.power .-= minimum(state.power[idx_finite]) # prevent subtracting -Inf
state.power[.!idx_finite] .= 0
return state
end
function ica_compete!(state::IcaState, optimizer::Ica)
if length(state.empire) == 1 ||
all(==(0), state.power) ||
rand() > optimizer.competition_pressure
return state
end
idx_src = argmin(state.power)
# sample empire index with given probabilities
idx_dst = findfirst(cumsum(state.power) .> rand() * sum(state.power))
# sample colony index uniformly
idx_col = rand(1:length(state.empire[idx_src]))
@debug "MOVE colony $(idx_col) (country $(state.empire[idx_src][idx_col])) from empire $(idx_src) to empire $(idx_dst)"
# move selected colony to destination empire
push!(state.empire[idx_dst], state.empire[idx_src][idx_col])
deleteat!(state.empire[idx_src], idx_col)
# merge source empire with strongest if only one colony remaining
if length(state.empire[idx_src]) <= 1 # can be 0 if an empire was initialized with just 1 colony
@debug "DELETE empire $(idx_src)"
# move last remaining colony to strongest empire
if length(state.empire[idx_src]) == 1
push!(state.empire[idx_dst], state.empire[idx_src][1])
end
# move former imperialist to strongest empire
push!(state.empire[idx_dst], state.imperialist[idx_src])
# delete the former empire and imperialist and its power
deleteat!(state.empire, idx_src)
deleteat!(state.imperialist, idx_src)
deleteat!(state.power, idx_src)
end
return state
endNotable differences to the ICA implementation in the ICAOD package are:
- no local search phase
- Monte-Carlo integration instead of HCubature
uniting_thresholdis an absolute quantity- collisions with the boundary are handled differently
Example
For illustration, we reuse the example with the discrete prior.
using Kirstine, Random, Plots
@simple_model SigEmax dose
@simple_parameter SigEmax e0 emax ed50 h
function Kirstine.jacobianmatrix!(
jm,
m::SigEmaxModel,
c::SigEmaxCovariate,
p::SigEmaxParameter,
)
dose_pow_h = c.dose^p.h
ed50_pow_h = p.ed50^p.h
A = dose_pow_h / (dose_pow_h + ed50_pow_h)
B = ed50_pow_h * p.emax / (dose_pow_h + ed50_pow_h)
jm[1, 1] = 1.0
jm[1, 2] = A
jm[1, 3] = -A * B * p.h / p.ed50
jm[1, 4] = c.dose == 0 ? 0.0 : A * B * log(c.dose / p.ed50)
return jm
end
prior = PriorSample(
[SigEmaxParameter(e0 = 1, emax = 2, ed50 = 0.4, h = h) for h in 1:4],
[0.1, 0.3, 0.4, 0.2],
)
dp = DesignProblem(
criterion = DCriterion(),
region = DesignInterval(:dose => (0, 1)),
model = SigEmaxModel(sigma = 1),
covariate_parameterization = CopyTo(:dose),
prior_knowledge = prior,
)
strategy = DirectMaximization(
optimizer = Ica(
iterations = 100,
num_countries = 100,
num_initial_empires = 20,
# settings (mostly) as in ICAOD defaults
revolution_rate = 0.3,
damp = 0.99,
zeta = 0.1,
beta = 4,
competition_pressure = 0.11,
uniting_threshold = 1.0,
),
prototype = equidistant_design(region(dp), 8),
)
Random.seed!(31415)
s, r = solve(dp, strategy, maxweight = 1e-3, maxdist = 1e-2, trace_state = true)
gd = plot_gateauxderivative(s, dp)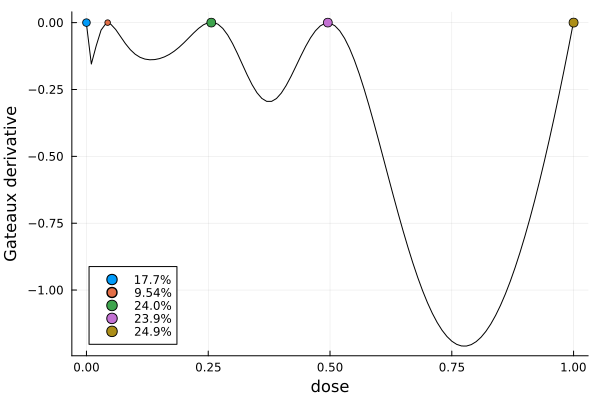
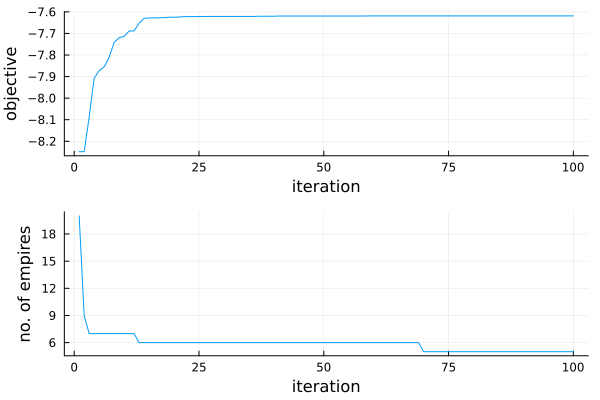
n_emp = map(st -> length(st.empire), optimization_result(r).trace_state)
dia = plot(
plot(r),
plot(n_emp; xguide = "iteration", yguide = "no. of empires", legend = nothing),
layout = (2, 1),
)